All-in-One™ miRNA qPCR Primers Sample Validation Report |
 |
- Materials and Methods
- Procedure
- Test Results
- Limited Warranty
|
 |
I. Materials and Methods |
- Instrument
iQ5 Real Time PCR Detection System: Bio-Rad
- Reagents
All-in-One™ miRNA qRT-PCR Detection Kit (Catalog Nos. AOMD-Q020 or AOMD-Q050) Validation templates: cDNAs extracted from 10 different human tissues Detection primers: Total 6 pairs
|
| Catalog number |
Primer ID |
Mature_Acc |
Mature_ID |
PCR size |
|
| miRQP0227 |
hsmq-0031 |
MIMAT0000069 |
hsa-miR-16 |
75bp |
| miRQP0074 |
hsmq-0032 |
MIMAT0000422 |
hsa-miR-124 |
73bp |
| miRQP0096 |
hsmq-0034 |
MIMAT0000423 |
hsa-miR-125b |
75bp |
| miRQP0175 |
hsmq-0035 |
MIMAT0000429 |
hsa-miR-137 |
76bp |
| miRQP9901 |
hsnRNA-U6 |
NR_002752.1 |
RNU6B |
80bp |
| MiRQP0293 |
hsmq-0655 |
MIMAT0000073 |
hsa-miR-19a |
72bp |
|
| |
II. Procedure
RNA Extraction
1. Sample preparation |
Place a small amount of material from a human tissue sample into a pre-cooled mortar. Add a small amount of liquid nitrogen and grind the tissue to a fine powder. Transfer the powder to a centrifuge tube containing 1 ml of TRIzol (Invitrogen). Vortex for 5 minutes. (If the samples are cultured cells, use about 106~107 cells with 1 ml of TRIzol). Pipette up and down until the cells are completely lysed).
|
2. Phase separation |
Leave the cell or tissue samples at room temperature for about 10 minutes. Add 200 μl of chloroform per 1 ml of TRIzol. Close the cap and vortex vigorously for 1 minute. Let the samples settle down at room temperature for 2-5 minutes. Centrifuge at 12,000 g for 15 minutes at 6°C. Remove tubes from the centrifuge being careful not to disturb the liquid. The samples should be separated into 3 layers with RNA in the top layer.
|
3. RNA Precipitation
|
Carefully transfer about 450 μl (per 1 ml TRIzol) of the supernatant to a new centrifuge tube containing 600 μl of cold 2-propanol. Mix well and keep at –20°C for 10 minutes. Then centrifuge at 12,000 g for 10 minutes (at 6°C).
|
4. Washing |
Remove the supernatant. Add 500 μl of cold 75% ethanol. Vortex until a pellet forms at the bottom of the tube. Centrifuge at 12,000 g for 5 minutes (6°C). Remove the supernatant. Centrifuge briefly again and remove the remaining supernatant.
|
5. Dissolve RNA |
Air-dry the samples for 5 to 10 minutes. (Do not over dry the samples. The samples are dry when they turn white). Dissolve the samples in 30 μl of DEPC water. Label the samples properly and store them at –80°C.
|
6. Determine RNA Concentration |
Take 1 μl of RNA sample. Dilute the sample 1:10 in DEPC water. RNA concentration can be measured with a NanoDrop (Thermo Scientific). DEPC water should be used as a blank. Record both the RNA concentration and the ratio of A260/A280.
|
7. RNA electrophoresis |
- Prepare denaturing gel Add 1 g of agarose into 75 ml of de-ionized water. Boil the agarose for 1–2 minutes and cool it down to about 70°C. Add 10 ml of 10X MOPS, 15 ml of formaldehyde and 5 μl of ethidium bromide (EB). Pour the gel into a tray with big combs. Cover the tray.
- Prepare electrophoresis buffer (1X MOPS) Take 50 ml of 10X MOPS and dilute 1:10 to the final volume of 500 ml with de-ionized water. Pour the buffer into a gel box, and add some EB to it.
- Preparation of RNA sample Take 3 μl of the RNA sample and add DEPC to a total volume of 18 μl. Heat the sample at 65°C for 10 minutes. Cool it down immediately. Add 2 μl of 1X RNA loading buffer into the sample
- RNA electrophoresis
Place the RNA gel in a gel box with 1X MOPS electrophoresis buffer. Pre-run the gel at 100 V for 5 minutes. Load the treated RNA samples into the gel and run at 100 V until the bromophenol blue runs to about one third the length of the gel. Take a picture using a UV scanner.
|
8. RNA detection results |
Electrophoresis results of RNAs from 10 different tissues(3 μl RNA each well). See table 8.2 for the specific lane information.
|
 |
| |
| Lane |
Tissue |
Concentration (ng/μl) |
OD A260/A280 |
|
| 1 |
Brain |
2385 |
1.9 |
| 2 |
Lung |
2430 |
1.94 |
| 3 |
Liver |
2521 |
1.91 |
| 4 |
Kidney |
3444 |
1.94 |
| 5 |
Breast |
2785 |
1.87 |
| 6 |
Testis |
2972 |
1.9 |
| 7 |
Placenta |
3515 |
1.91 |
| 8 |
Spleen |
3344 |
1.91 |
| 9 |
Heart |
3394 |
1.91 |
| 10 |
Pancreas |
3101 |
1.91 |
|
| |
| Note: In order to detect miRNAs, the extracted RNA must contain small molecular weight RNAs. The kit used for RNA extraction must be applicable for isolation of total RNA or small RNA. The quality of RNA is critical to the success of downstream experiments. Follow the instructions of the RNA extraction kit exactly. Examine RNA samples by electrophoresis to ensure high quality. |
| |
miRNA reverse transcription |
- Thaw all the reagents needed for miRNA reverse transcription. Mix reagents well by gently inverting the tubes. Spin down briefly and keep on ice.
- Prepare miRNA reverse transcription reaction: Add the following reagents into an RNase-free reaction tube which is pre-cooled on ice. The final volume should be 25 μl.
|
| Reagents |
Volume |
Final concentration |
|
| Total RNA |
Brain |
2 μg |
| 2.5 U/μl Poly A Polymerase |
1 μl |
2.5 U |
| RTase Mix |
1 μl |
2521 |
| 5X Reaction Buffer |
5 μl |
1X |
| ddH2O (RNase/DNase free) |
to 25 μl |
|
|
| Note: The total RNA in the reaction must contain small molecular RNA. If total RNA is used, the amount of RNA should be between 1 ng ~ 5 μg. If the purified small molecular RNA is used, the amount of RNA should be between 0.1 ng ~ 1 μg |
| |
- Reverse Transcription Reaction: Mix reaction solution well. Spin down briefly. Incubate the reaction solution at 37°C for 60 minutes. Terminate the reaction by heating at 85°C for 5 minutes. The products of reverse transcription can be diluted 1:5 with sterile water for the downstream qPCR reaction.
|
| |
qPCR to detect miRNA |
- Thaw 2X All-in-One qPCR mix from the All-in-One miRNA qRT-PCR Detection Kit. Mix well by inverting the tube several times. Spin down briefly and keep on ice.
- Prepare qPCR reaction solution on ice. All miRNAs are tested in duplicate. NTC (No template control) is tested singly.
|
| Reagents |
Volume |
Final concentration |
|
| 2×All-in-One qPCR Mix |
10 μL |
1X |
| All-in-One miRNA qPCR Primer |
(2 μM) 2 μL |
0.2 μM |
| Universal Adaptor PCR Primer |
(2 μM) 2 μL |
0.2 μM |
| First strand cDNA (diluted 1:5) |
2 μL |
|
| ddH2O |
4 μL |
|
| Final volume |
20 μL |
|
|
| |
- Mix the qPCR reaction solution well. Transfer the solution to a PCR tube. Spin briefly to make sure that the reaction solution is at the bottom of the tube
- The qPCR reaction, using the standard 3-step method to detect the DNA amount (these experiments were designed based on Bio-Rad iQ5 instrument). Melting analysis can be done immediately after the amplification reactions.
|
| Number of Cycles |
Step |
Temperature |
Time |
Detection |
|
| 1 |
Pre-denature |
95 °C |
10min |
No |
| 40 |
Denature |
95 °C |
10sec |
No |
| 40 |
Annealing |
See reference below |
20sec |
No |
| 40 |
Extension |
72 °C |
10sec |
Yes |
|
| |
| |
| Temperature range |
Rate of temperature change |
Duration |
Detection |
|
| 66 °C ~ 95 °C |
0.5 °C/ step |
6 sec/ step |
Yes |
| 30 °C |
|
30 sec |
No |
|
| |
| Note: The above conditions are designed for use with the Bio-Rad iQ5Q-PCR instrument. If other instruments are used, adjust the extension time and conditions for analysis as recommended by the manufacturers. The annealing conditions for each miRNA assay may be different. Please refer to the details of the conditions in the following section. |
| |
III. Test Results
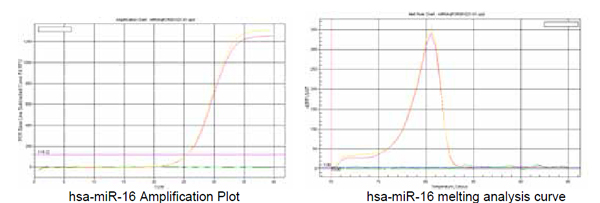
1. hsa-miR-16(hsmq-0031 primer)
Recommended annealing temperature:60℃
|
 |
| |
2. hsa-miR-124(hsmq-0032 primer) |
Recommended annealing temperature:60℃
|
 |
| |
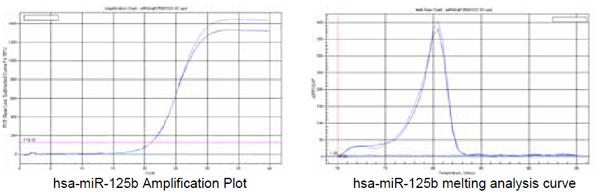
3. hsa-miR-125b (hsmq-0034 primer) |
Recommended annealing temperature:60℃
|
 |
| |
4. hsa-miR-137 (hsmq-0035 primer) |
Recommended annealing temperature:60℃
|
 |
| |
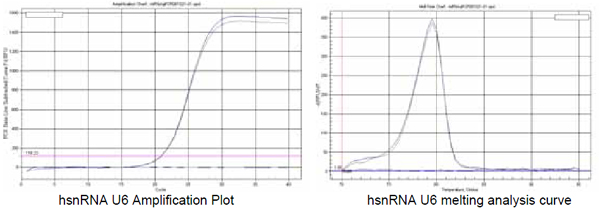
5. hsnRNA U 6 primer testing |
Recommended annealing temperature:60℃
|
 |
| |
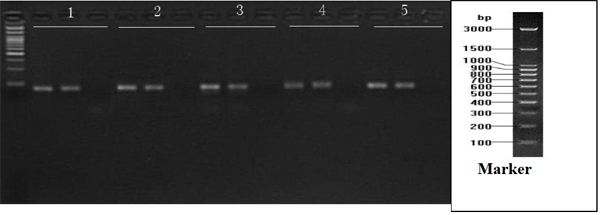
6. Electrophoresis Results
|
 |
| |
5 μl of the PCR of products, run on 3% agarose gel. Each set contains two positive controls and one NTC Lane 1: hsa-miR-16; Lane 2: hsa-miR-124; Lane 3: hsa-miR-125b; Lane 4: hsa-miR-137; Lane 5: hsnRNA-U6.
|
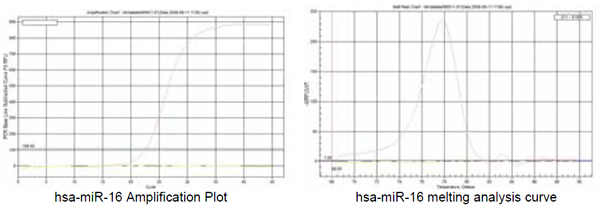
7. hsa-miR-19a(hsmq-0655 primer)
7.1 Validation. |
The following link may be used to view a larger amplification plot and melting curve as well as a gel electrophoresis image of this primer’s validation results. Contact GeneCopoeia at inquiry@genecopoeia.com or call 866-360-9531 to have an electronic document sent to you.
|
 |
| |
7.2 Discrimination assay of two primers with single-base differences in their sequences |
| 7.2.1 has-miR-19a BLAST |
| hsa-miR-19a |
UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b |
UGUGCAAAUCCAUGCAAAACUGA |
|
| |
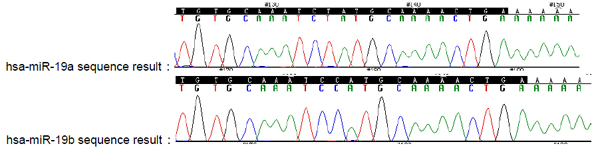
| The BLAST alignment shows that a single-base difference exists between the two miRNAs: hsa-miR-19a and miR-19b. To discriminate and validate the primer of interest (has-miR-19a) two plasmids were constructed for each of the two miRNAs. |
| |
7.2.2 Sequence result of the miRNA qPCR plasmids |
 |
| |
7.2.3 qPCR validation |
Using the All-in-One miRNA qRT-PCR Detection Kit, the hsa-miR-19a-specific primer was validated using miRNA expression plasmids for has-miR-19a and has-miR-19b as templates (about 107 molecules/reaction).
The percent relative discrimination was calculated using the differences between the Ct values of the mismatched template and the matching template with respect to the hsa-miR-19a-specific primer (Relative discrimination =2-ΔCt ×100%). The results are shown in the table below: |
| Annealing temperature °C |
hsa-miR-19a |
hsa-miR-19b |
hsa-miR-19b |
|
| 63.3 |
20.30 |
30.89 |
0.0651% |
| 61.4 |
18.40 |
25.81 |
0.5880% |
| 58.9 |
17.50 |
21.57 |
5.9545% |
|
| |
7.2.4 Conclusion |
Based on these results, a qPCR annealing temperature between 59.9 and 61.4 °C is recommended. This range gives the highest amplification efficiency as well as the best relative discrimination between the two single-base-different miRNAs.
|
IV. Limited Use License and Warranty
Limited Use License |
Following terms and conditions apply to use of all OmicsLink™ ORF Expression Clones in all lentiviral vectors and Packaging Kit (the Product). If the terms and conditions are not acceptable, the Product in its entirety must be returned to GeneCopoeia within 5 calendar days. A limited End-User license is granted to the purchaser of the Product. The Product shall be used by the purchaser for internal research purposes only. The Product is expressly not designed, intended, or warranted for use in humans or for therapeutic or diagnostic use. The Product must not be resold, repackaged or modified for resale, or used to manufacture commercial products without prior written consent from GeneCopoeia. This Product should be used in accordance with the NIH guidelines developed for recombinant DNA and genetic research. Use of any part of the Product constitutes acceptance of the above terms.
|
Limited Warranty |
GeneCopoeia warrants that the Product meets the specifications described in the accompanying Product Datasheet. If it is proven to the satisfaction of GeneCopoeia that the Product fails to meet these specifications, GeneCopoeia will replace the Product. In the event a replacement cannot be provided, GeneCopoeia will provide the purchaser with a refund. This limited warranty shall not extend to anyone other than the original purchaser of the Product. Notice of nonconforming products must be made to GeneCopoeia within 30 days of receipt of the Product. GeneCopoeia’s liability is expressly limited to replacement of Product or a refund limited to the actual purchase price. GeneCopoeia’s liability does not extend to any damages arising from use or improper use of the Product, or losses associated with the use of additional materials or reagents. This limited warranty is the sole and exclusive warranty. GeneCopoeia does not provide any other warranties of any kind, expressed or implied, including the merchantability or fitness of the Product for a particular purpose.
GeneCopoeia is committed to providing our customers with high-quality products. If you should have any questions or concerns about any GeneCopoeia products, please contact us at 301-762-0888. |
